Deep learning solves a 20-year long unsolved problem in science (Part 1)
DeepMind changes drug discovery and genetic research forever
In previous iterations of this newsletter, we’ve discussed deep learning primarily through the lends of creative avenues like text, code and image generation. This is because deep learning is particularly good at creating an efficient (if lossy) representation of a vast amount of information. This gives us the illusion of creativity, and thus makes models excel.
Ted Chiang in the New Yorker calls ChatGPT “a blurry jpeg of the web” — a compression of a lot of data. This compression power is what intuitively might make large language models great in data-rich scientific domains. A library of 1000 genomes takes up >200Tb of data. One can only imagine how intensive it is to actually utilize scientific datasets of this scale.
This is a problem specifically in protein folding. Proteins are chains of amino acids. The specific configuration of the chain and their subsequent interactions create unique 3D structures.
These structures determine specific properties of said protein — learning about them can aid research and drug discovery. But, as one can imagine, they are hard to map — it might take months to years to add a single protein’s structures.
That would be the case, of course, until deep learning changed everything. Today, we talk about “Highly accurate protein structure prediction for the human proteome with AlphaFold” by DeepMind.
Wait, “Part 1”?
A quick clarification — yes, this is part one of two. This is because AlphaFold might be one of the most important papers of the last couple of years. It was the most cited paper of 2021 and 2022. It has cleaved the world of computational biology in “before AlphaFold” and “after AlphaFold”.
And it’s dense. This paper is a masterclass in how to tailor deep learning to a specific use case. It’s just going to take more than one post to wring out all the insights from this one.
Introduction and Motivation
Goal: Given an amino acid sequence, what would their interactions and the subsequent protein structure look like?
Constraints: A first approach might suggest doing what we’ve done with most ML problems — create a dataset that maps amino acid sequence → 3D protein structure, train until useful. The big problem — deep learning is quite data-hungry and there only exist 100,000 such mappings in the world. The computation cost would be unreasonable.
In the words of the author, “it is not normal to do machine learning in a domain where every data point represents a PhD’s worth of effort”.
Solution: How do we proceed? We think about why humans don’t face the same problems — it’s because we understand insights from physics and geometry, that we leverage to build 3D structures faster. These inductive biases are what the authors attempt to give to the model to help it learn from a smaller dataset.
Development Details
Let’s discuss AlphaFold’s prediction structure at a very high level. Strap in, this one’s a doozy.
We first realize that the complex spatial geometry of proteins cannot be captured by standard nets like CNNs or RNNs. Thus, it is essential to use an attention model, which can make data-driven decisions about the extent to which amino acids interact with each other. (This is the power of all attention models, btw).
But even with using attention, the prediction model is missing the aforementioned “inductive biases” — notion of geometry and the like. Thus, we give the model these biases through the first step: data-fetching. Given an input sequence,
we pull “multi-sequence alignments” — the evolutionary history of a particular sequence, from a database.
We also pull “template structures” of similar sequences from another database.
Finally, we build out a blank system of “pairwise residue terms”. (Residue is a particular amino acid in the chain). You feed these into the network as well. This encodes our expectation of pairwise interaction between residue in nature.
We feed all three of these into our second step: the Evo-former. This is the primary deep learning model in the system. It inputs an MSA and the pairwise terms, and outputs a refined version of the MSA. There are many techniques used here to improve the inductive bias of the model.
Finally, the outputs of this are fed into the third step: the Structure Module. This module is defined such that it understands the 3D structure and physics of the refined MSA. It is meant to output a set of attributes equivalent to the 3D structure of the protein.
The final idea I’d mention is that of “recycling”. Since they are training their model to refine the MSA, one can take a refined MSA and treat it as the input again, creating a chain of refinement.
Evaluation
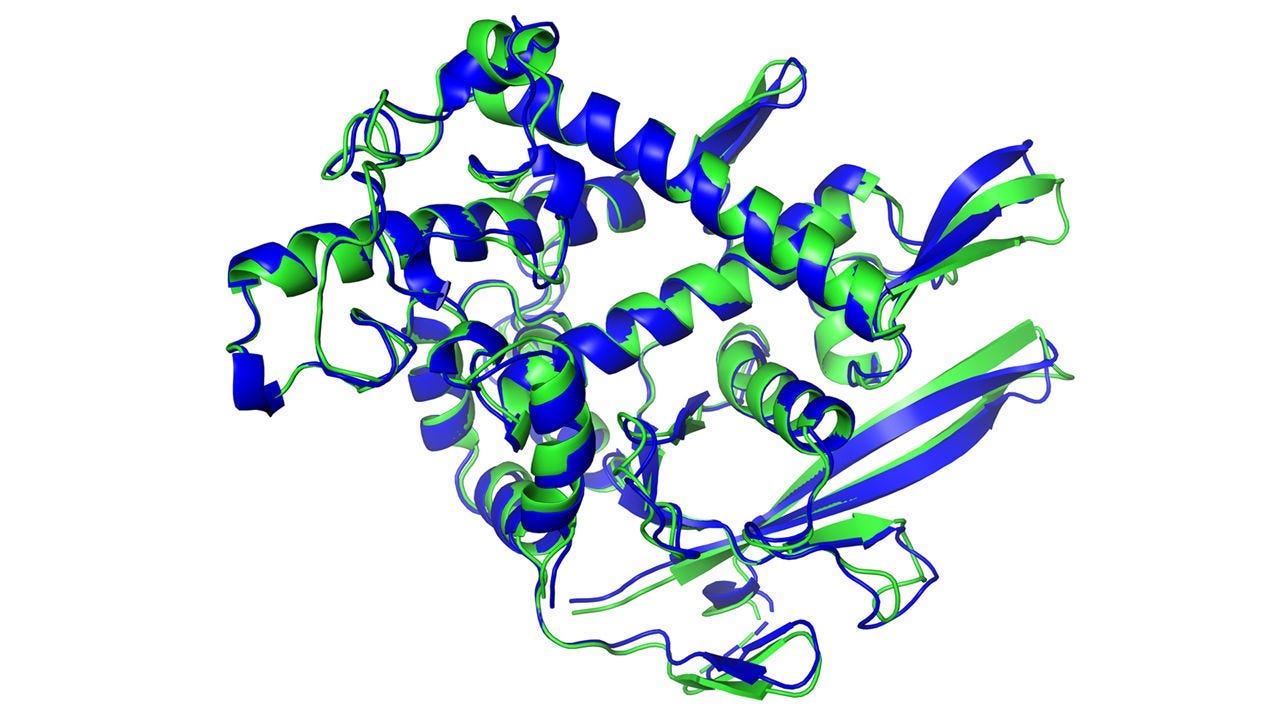
We first look at a comparison of AlphaFold’s predictions vs. a test set of pdb chains. The green is the “ground truth”, the blue is AlphaFold. Note how close AlphaFold’s predictions are.
If you’re like me, you have no idea if this data is good or bad. Let me add context — the predictions are close enough to predict the function and properties of the proteins correctly, thus acting as a decent proxy for the actual structure.
Second, we look at a comparison of the median root mean squared deviation between the true and predicted structure. As you can see, AlphaFold blows its competitors out of the water.
Midpoint
An evaluation of such an intricate model would be incomplete without some ablation studies, and ablation studies are pointless without understanding some model details first. We’ll get to this next time.
For now, I’d like to end off this section with some context. The human genome was first completely mapped in 2003. This ushered in an era of “sequence abundance”, leading to scientific advancements, better treatment of diseases and genetic disorders, a better understanding of human evolution, and more.
AlphaFold ushers in an era of “structure abundance”. Who is to say what might become possible in the next decade?
Until next time!