Deep learning solves a 20-year long unsolved problem in science (Part 2)
Deepmind changes drug discovery and genetic research forever
Last Time
We took a look at “AlphaFold”, the deep learning model by DeepMind that creates a mapping from a sequence of amino acids to 3D protein structure. This is critical in analyzing the functioning and properties of proteins, as well as drug discovery research. Check out part 1 here and then jump into this one!
We looked at some cool data, and developed a preliminary understanding of the model structure:
We talked about how AlphaFold was uniquely powerful in that the model architecture was tailored for the problem of protein structures, by adding inductive biases to the model. Today, we’ll talk about model details to get some intuition about how to develop some models ourselves and do some ablation evaluations!
Model Details
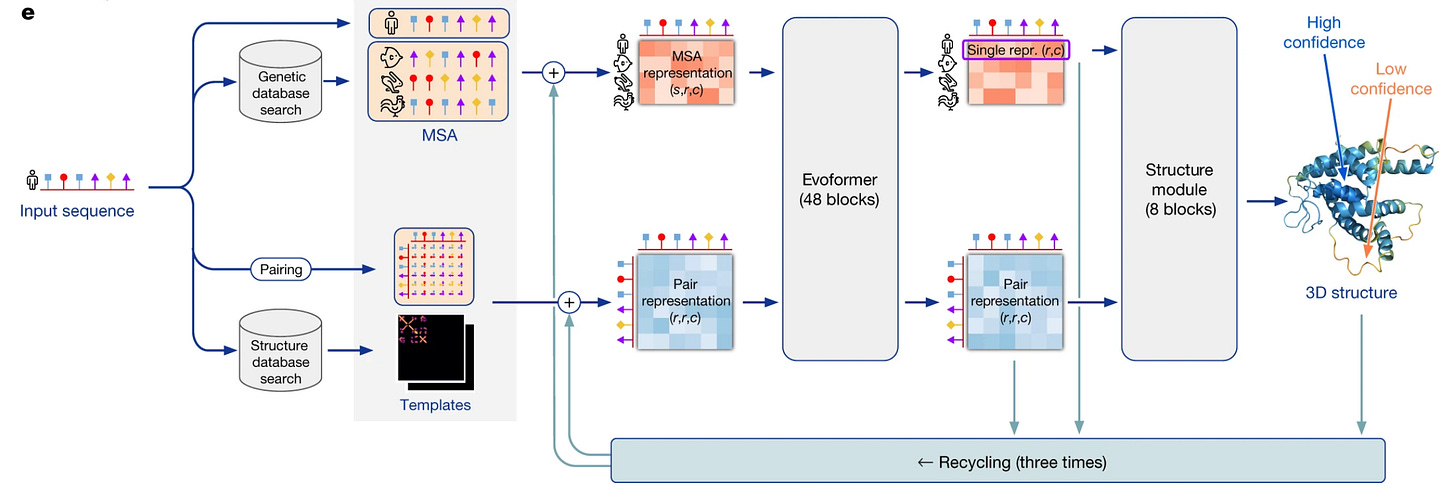
We’ll split this section into two parts: talking about Evoformer and talking about the Structure Module. In talking about these models, we’ll attempt to keep the focus on why these models look like what they look like.
Evoformer
The evoformer forms the base of the model architecture. The core purpose of the evoformer is to map a set of protein structures to 1 protein structure.
What is the input?
MSA + Residue Pair Representation.
MSA: multi-sequence alignment, pulling similar sequences to the query from a database
Residue Pair Representation: an interaction matrix between residue pairs
The residue pairs encode spatial relationships, the MSA encodes evolutionary knowledge
Proteins co-evolve → their evolutionary history determines their structural correlations as well.
How do the two communicate?
MSA + Residue pairs communicate through two main avenues — pair bias in attention, and outer product mean.
The bias term is added to attention. Standard attention looks like
\((QK^T)V \)whereas the bias term in attention added looks like
\((QK^T + b)V\)This bias term conveys some prior hypothesis regarding the distance between element i and j. These new hypotheses developed are tested.
Outer product mean is used to combine the MSA and residue pair matrices together. The reason this is picked over other operations is because an outer product gives a combination of features of both matrices, and the mean creates average features that combine the two.
How do we encode physical relationships in the model itself?
The big difference between generating 3D models and generating GPT-3 style text is that 3D models must work according to the rules of physics.
One can think of the distances matrix (the residue pairs) as an adjacency matrix.
We must care about not only interactions between i and j, but also the combination between i, j, and k.
A simple representation of this is the triangle inequality, which all objects must follow in nature.
Thus, using a 3-way update for attention instead of just 2 ways, we encode this simple physics law in our system.
Structure Module
Once the data is run through the Evoformer, we need to convert it to a format that can be compared to the 3D structures we have during training. The Evoformer converts the sequence to a 1D embedding of a protein structure. We use structure module to convert the 1D embedding into a 3D structure.
At a very high level, this block takes a list of relative positions (amino acid A is to the south of B, B is to the west of C…) and puts them together in a 3D structure, then tunes their relative rotations and translations.
One of the most interesting components is the IPA — the invariant point attention. Without going into too much detail, this attention in invariant to global rotation and translation. This means that we can extract the maximal amount of information when comparing to our ground truth. This block is particularly interesting, since it is primarily for faster learning during training. Remember: we have very limited data — we have to use it as best we can!
Recycling
The core intuition behind recycling is to re-send the outputs of the model through the model such that it allows the network to further refine structures which is it less confident about, by some intrinsic accuracy estimation.
Very literally, the model is “refining” the 3D structures it is creating!
Evaluation
Wooooow that was a lot of technical detail. Hopefully, they gave you some insight into what model development looks like.
But a model is only as good as every component of it is useful. Let’s look at some ablation data! The graph below demonstrates some model architecture details we talked about, and many we didn’t.
The core observation: Everything contributes something to the model accuracy.
Here’s another really interesting observation — sometimes, the model will generate 3D structure with “boring” sections of thin ribbons. There is a correlation between model confidence estimation being very low and the boring section. If the model is uncertain, that’s a strong signal of disorder or an unstructured chain.
This seems to imply that the model is learning something after all!
In Summary
Now you probably know more about AlphaFold than you ever wanted to. There’s a couple big ideas I want to highlight here:
Training models is an art — AlphaFold uses a database for initialization, uses physics laws to train faster, and uses principles of invariance in nature to make the loss function more accurate. You can’t “just” train a model. You need to be really thoughtful about what your model is doing, and how to best create emergent phenomena.
Using just the transformer isn’t enough — I can’t tell you how many papers I’ve read that stick to the vanilla Transformer model. AlphaFold is proof that being more creative with your neural net modules will create massively better performance on specific tasks.
Low data regimes — most problems in the world are low on data. If we hope to build neural solutions for these, techniques for creating models out of less data is essential to growth.
AlphaFold really is a beautiful paper, a one of a kind project in the land of quick paper turn-arounds. I’d really recommend giving it a go yourself.
I know these two editions were long and quite technical, so thank you for sticking through them with me! Next week, we’ll be back to our regularly scheduled programming, and I hope you’re excited 4 it 😉
Until next time!